Chromatography
Column chromatography is one of the most common methods of protein purification. Like many of the techniques on this site, it is as much an art form as a science. Proteins vary hugely in their properties, and the different types of column chromatography allow you to exploit those differences. Most of these methods do not require the denaturing of proteins.
To be very general, a protein is passed through a column that is designed to trap or slow up the passing of proteins based on a particular property (such as size, charge, or composition).
There are three main steps to protein purification:
1. Capture. You need to get your protein into a concentrated form. If, for example, you are trying to isolate a protein you have synthesized in an E. coli cell, you could be looking at a protein to junk ratio of 1:1,000,000. For capture purification you need a high capacity method that is also fast. You need a speedy method because your crude solution is very likely to also contain proteases and these can quickly chew up your protein.
2. Intermediate. Intermediate purification requires both speed and good resolution.
3. Polishing. For the final step of purification you need a system that has both good resolution and speed. Capacity is usually irrelevant at this stage.
Some of the more common columns include:
IEX: Ion exchange chromatography. Good for capture, intermediate, and polish.
HIC: Hydrophobic interaction column. Good for intermediate purification.
AC: Affinity chromatography. Good for capture and intermediate purification.
GF: Gel filtration (size exclusion) chromatography. Good polishing step.
Let's look at these types of columns in more detail.
Ion exchange chromatography
Ion exchange chromatography is based on the charge of the protein you are trying to isolate. If your protein has a high positive charge, you'll want to pass it through a column with a negative charge. The negative charge on the column will bind the positively charged protein, and other proteins will pass through the column. You then use a procedure called "salting out" to release your positively charged protein from the negatively charged column. The column that does this is called a cation exchange column and often uses sulfonated residues. Likewise, you can bind a negatively charged protein to a positively charge column. The column that does this is called an anion exchange column and often uses quaternary ammonium residues.
Salting out will release, or elute, your protein from the column. This technique uses a high salt concentration solution. The salt solution will out-compete the protein in binding to the column. In other words, the column has a higher attraction for the charge of salts than for the charged protein, and it will release the protein in favor of binding the salts instead. Proteins with weaker ionic interactions will elute at a lower salt, so you will often want to elute with a salt gradient. Different proteins elute at different salt concentrations, so you will want to be sure you know well the properties of your protein best results.
Also be aware that changes in pH alter the charges in proteins. Be sure you know the isoelectric point of your protein (the isoelectric point is the pH at which the charge of a protein is zero) and make sure the pH of your system is adjusted and buffered accordingly.
The basic steps in using an ion exchange column are:
1. Prep the column. Pour your buffer over the column to make sure it has equilibrated to the required pH.
2. Load your protein solution. Some proteins in the solution don't bind and will elute during this loading phase.
3. Salt out. Increase the salt concentration to elute the bound proteins. It is best to use a salt gradient to gradually elute proteins with different ionic strengths. At the end bump the system with a very high salt concentration (2-3M) to make sure all proteins are off the column.
4. Remove salts. Use dialysis to remove the salts from your protein solution.
Temperature doesn't have a huge effect on column chemistry. However, it is better to work cold since proteins are more stable cold.
Hydrophobic interaction chromatography
Where ion exchange chromatography relies on the charges of proteins to isolate them, hydrophobic interaction chromatography uses the hydrophobic properties of some proteins. Hydrophobic groups on the protein bind to hydrophobic groups on the column. The more hydrophobic a protein is, the stronger it will bind to the column.
Load the proteins in the presence of a high concentration of ammonium sulfate (not ammonium persulfate). Ammonium sulfate is a chaotropic agent. It increases the chaos (entropy) in water, and thereby increases hydrophobic interactions (the more disordered the water, the stronger the hydrophobic interactions). Ammonium sulfate also stabilizes proteins. So as a result of using an HIC column you can expect your protein to be in its most stable form.
The hydrophobic column is packed with a phenyl agarose matrix. In the presence of high salt concentrations the phenyl groups on this matrix binds hydrophobic portions of proteins. You can control elution of different column-bound proteins by reducing the salt concentration or by adding solvents.
Affinity chromatography.
 Affinity chromatography relies on the biological functions of a protein to bind it to a column. The most common type involves a ligand, a specific small biomolecule. This small molecule is immobilized and attached to a column matrix, such as cellulose or polyacrylamide. Your target protein is then passed through the column and bound to it by its ligand, while other proteins elute out. Elution of your target protein is usually done by passing through the column a solution that has in it a high concentration of free ligand. This is a very efficient purification method since it relies on the biological specificity of your target protein, such as the affinity of an enzyme for a substrate.
Affinity chromatography relies on the biological functions of a protein to bind it to a column. The most common type involves a ligand, a specific small biomolecule. This small molecule is immobilized and attached to a column matrix, such as cellulose or polyacrylamide. Your target protein is then passed through the column and bound to it by its ligand, while other proteins elute out. Elution of your target protein is usually done by passing through the column a solution that has in it a high concentration of free ligand. This is a very efficient purification method since it relies on the biological specificity of your target protein, such as the affinity of an enzyme for a substrate.
Gel filtration (size exclusion) chromatography d
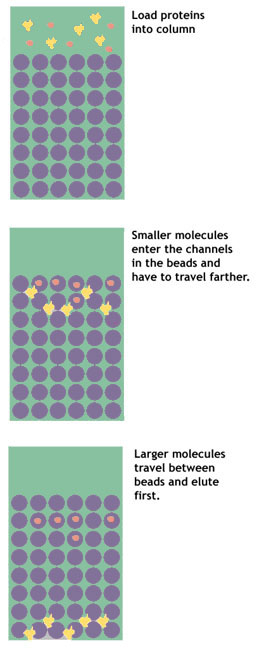 Gel filtration, or size exclusion, chromatography separates proteins on the basis of their size. The column is packed with a matrix of fine porous beads.
Gel filtration, or size exclusion, chromatography separates proteins on the basis of their size. The column is packed with a matrix of fine porous beads.
It works somewhat like a sieve, but in reverse. The beads have in them very small holes. As the protein solution is poured on the column, small molecules enter the pores in the beads. Larger molecules are excluded from the holes, and pass quickly between the beads.
These larger molecules are eluted first. The smaller molecules have a longer path to travel, as they get stuck over and over again in the maze of pores running from bead to bead. These smaller molecules, therefore, take longer to make their way through the column and are eluted last.
Read more about: Mama Ji's Molecular Kitchen
Bibliographic details:
- Article: Chromatography
- Author(s): Dr. Biology
- Publisher: Arizona State University School of Life Sciences Ask A Biologist
- Site name: ASU - Ask A Biologist
- Date published: 12 Apr, 2010
- Date accessed:
- Link: https://askabiologist.asu.edu/chromatography
APA Style
Dr. Biology. (Mon, 04/12/2010 - 14:36). Chromatography. ASU - Ask A Biologist. Retrieved from https://askabiologist.asu.edu/chromatography
Chicago Manual of Style
Dr. Biology. "Chromatography". ASU - Ask A Biologist. 12 Apr 2010. https://askabiologist.asu.edu/chromatography
Dr. Biology. "Chromatography". ASU - Ask A Biologist. 12 Apr 2010. ASU - Ask A Biologist, Web. https://askabiologist.asu.edu/chromatography
MLA 2017 Style
Be Part of
Ask A Biologist
By volunteering, or simply sending us feedback on the site. Scientists, teachers, writers, illustrators, and translators are all important to the program. If you are interested in helping with the website we have a Volunteers page to get the process started.